Structural understanding of non-nucleoside inhibition in an elongating herpesvirus polymerase.
Hayes, R.P., Heo, M.R., Mason, M., Reid, J., Burlein, C., Armacost, K.A., Tellers, D.M., Raheem, I., Shaw, A.W., Murray, E., McKenna, P.M., Abeywickrema, P., Sharma, S., Soisson, S.M., Klein, D.(2021) Nat Commun 12: 3040-3040
- PubMed: 34031403
- DOI: https://doi.org/10.1038/s41467-021-23312-8
- Primary Citation of Related Structures:
7LUF - PubMed Abstract:
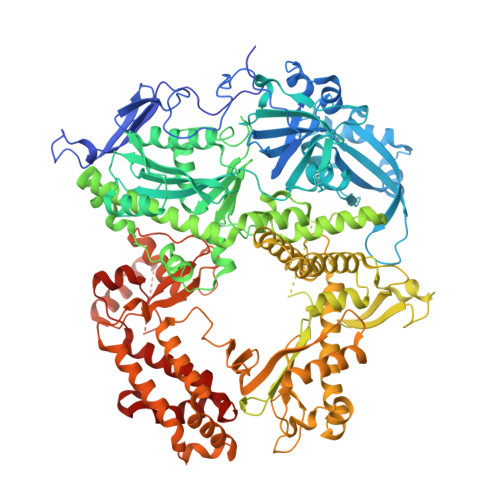
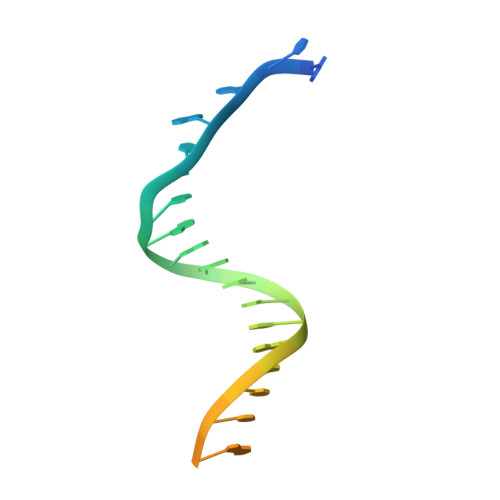
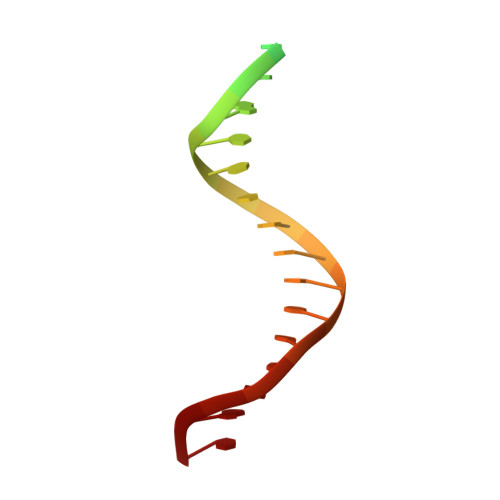
All herpesviruses encode a conserved DNA polymerase that is required for viral genome replication and serves as an important therapeutic target. Currently available herpesvirus therapies include nucleoside and non-nucleoside inhibitors (NNI) that target the DNA-bound state of herpesvirus polymerase and block replication. Here we report the ternary complex crystal structure of Herpes Simplex Virus 1 DNA polymerase bound to DNA and a 4-oxo-dihydroquinoline NNI, PNU-183792 (PNU), at 3.5 Å resolution. PNU bound at the polymerase active site, displacing the template strand and inducing a conformational shift of the fingers domain into an open state. These results demonstrate that PNU inhibits replication by blocking association of dNTP and stalling the enzyme in a catalytically incompetent conformation, ultimately acting as a nucleotide competing inhibitor (NCI). Sequence conservation of the NCI binding pocket further explains broad-spectrum activity while a direct interaction between PNU and residue V823 rationalizes why mutations at this position result in loss of inhibition.
- Computational and Structural Chemistry, Merck & Co., Inc., West Point, PA, USA. robert.hayes@merck.com.
Organizational Affiliation: